New publication from the Hopfner lab: "Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50"
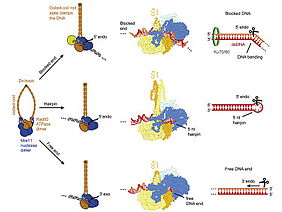
DNA double-strand breaks (DSBs) threaten genome stability and are linked to tumorigenesis in humans. Repair of DSBs requires the removal of attached proteins and hairpins through a poorly understood but physiologically critical endonuclease activity by the Mre11-Rad50 complex. Here, we report cryoelectron microscopy (cryo-EM) structures of the bacterial Mre11-Rad50 homolog SbcCD bound to a protein-blocked DNA end and a DNA hairpin. The structures reveal that Mre11-Rad50 bends internal DNA for endonucleolytic cleavage and show how internal DNA, DNA ends, and hairpins are processed through a similar ATP-regulated conformational state. Furthermore, Mre11-Rad50 is loaded onto blocked DNA ends with Mre11 pointing away from the block, explaining the distinct biochemistries of 3′ → 5′ exonucleolytic and endonucleolytic incision through the way Mre11-Rad50 interacts with diverse DNA ends. In summary, our results unify Mre11-Rad50’s enigmatic nuclease diversity within a single structural framework and reveal how blocked DNA ends and hairpins are processed.
Gut F, Käshammer L, Lammens K, Bartho JD, Boggusch AM, van de Logt E, Kessler B, Hopfner KP (2022) Structural mechanism of endonucleolytic processing of blocked DNA ends and hairpins by Mre11-Rad50. Mol Cell 12(22):715-718, https://doi.org/10.1016/j.molcel.2022.07.019